A BIOCHEMICAL STUDY ON ß-GLUCAN SYNTHESIS IN THE COTTON FIBER
Heungsop Shin and R. Malcolm Brown, Jr.Department of Botany, The University of Texas at Austin, Austin, Tx., 78713
ABSTRACT
Recently, our group achieved in vitro synthesis of the cellulose I allomorph (Plant Physiology 107:111,1995) from plasma membrane extracts of the cotton (Gossypium hirsutum) fiber. This success of the first in vitro cellulose I synthesis from higher plant cells was due to modification of the extraction and solubilization protocols of plasma membrane proteins. Under these improved conditions, 0.05 % digitonin solubilized fraction (SE1) synthesized a significant amount of cellulose I, whereas 1 % digitonin solubilized fraction (SE2) produced mostly b-1,3 glucan. Continuing this study, a product entrapment combined with photoaffinity labeling using azido-UDP[P32]-glucose has been used to identify and characterize polypeptides involved in b-glucan synthesis in the cotton fiber. Protein quantity and total b-glucan synthase activities in protein fractions released after product entrapment of SE1 and SE2 were prominently decreased; however, specific activities of b-glucan synthase were increased more than 4-fold. Two polypeptides, 34 and 35 kD, which were comparatively more prevalent in the SE1 fraction, were enriched after SE1 entrapment. Protein quantity of entrapped 34 and 35 kD polypeptides was affected largely by Ca2+ and Mg2+ concentrations in the product entrapment reactions. b-Glucan synthase activities from entrapped proteins corrrelated positively with the quantity of 34 and 35 kD polypeptides. Amino acid analysis of 34 and 35 kD polypeptides showed that they have a conserved sequence domain identical to plant annexins, implying Ca2+-regulated membrane binding properties of 34 and 35 kD polypeptides in vivo. Photoaffinity labeling experiments showed that 34 and 35 kD polypeptides could bind azido-UDP[P32]-glucose in vitro. These results suggest that the in vivo functions of 34 and 35 kD polypeptides could be important regulators of cellulose and callose synthesis in the cotton fiber.
INTRODUCTION
Cellulose, a b-1,4-glucan comprised of a single monomer and linkage type, is a primary component of the cell walls of higher plants. The process of cellulose biosynthesis in higher plants has still remained one of the major unresolved problems in plant biology.
As a first step to understand the mechanism of cellulose biosynthesis in higher plant cells, we have been studying, identifying, and isolating specific polypeptides which might be responsible for the cellulose synthesis in the cotton fiber. In the course of this work, we isolated two polypeptides from cotton fiber membranes, 34 and 35 kD, which were found to be prevalent in the cellulose-synthesizing enzyme fraction (SE1). Amino acid sequence analysis of the 34 and 35 kD polypeptides showed that they have a conserved sequence domain homologous to annexins, which is a family of Ca2+-binding proteins that bind to membranes in a Ca2+-dependent manner1,2. The annexins share a repetitive 17-amino acid sequence termed the endonexin fold3 which is contained within each of four domains each of which consists of 70 to 80 amino acids. A part of each domain is a Ca2+- and phospholipid-binding region. At present there is no known common function among the annexins.
Annexin-like proteins have been identified from other plants4,5,6 mostly based on amino acid sequence and functional similarities with animal annexins. Like the animal annexins, the functional roles for plant annexins are diverse. A recently identified annexin-like protein in cotton fibers7 was suggested to be involved in the inhibition of callose synthase activity in vitro; however, the exact functional role for cotton annexin-like protein in the regulation of callose synthesis is still obscure. In our study, cotton annexin-like proteins appear to positively correlate with callose synthase activities in vitro. In this poster, we present the biochemical properties and potential functions of cotton annexin-like proteins in b-glucan synthesis which are primarily based on the product entrapment8 and photoaffinity labeling experiments9.
We would also like to present our recent data of the identification of b-glucan synthase activities using native gel electrophoresis10. We have applied electron microscopic techniques11 to analyze the in vitro b-glucan products synthesized by the proteins that have been separated in native gels. Preliminary TEM product analysis data will be presented and discussed in this poster.
MATERIALS AND METHODS
Preparation of Membrane Proteins The cotton strain, Gossypium hirsutum Texas marker 1 was used for the experiments. The membrane proteins from cotton fibers were prepared by using the extraction buffer consisting of 50 mM Mops, pH 7.5, 5 mM EDTA, 0.25 M sucrose, and a combination of protease inhibitors. Generally, solubilized enzyme fractions were obtained separately through a two-step digitonin solubilization12. In the first step, a final concentration of 0.05 % digitonin was used. In the second step, a final 0.5 % or 1 % of digitonin was used.
Product Entrapment and Photoaffinity Labeling Further purification of the solubilized enzyme fractions was conducted by product entrapment. The reaction conditions for product entrapment were basically same as the conditions for the in vitro cellulose synthesis reaction. After entrapment reactions, the entrapped proteins with b-glucan products were collected by centrifugation, and the pellets were resuspended in buffer to release the entrappped proteins into the supernatant. The supernatant was designated as the ES fraction and was used for SDS-PAGE and activity assays. Azido-UDP[P32]-glucose was used as the substrate instead of UDP-glucose for photoaffinity labeling experiments9.
Isolation and Protein Sequencing of Annexins Bands of annexins were separated on SDS-PAGE gels after product entrapment reactions. The separated bands were digested with V8 proteases using 'in-gel' digestion methods13 and subjected to protein sequencing according to Matsudaira14.
Native Gel Electrophoresis A discontinuous gel system was used in performing native gel electrophoresis. 6 % of acrylamide was used for the separating gel and 4 % of acrylamide was used for the stacking gel. For detection of in situ enzyme activity, gels were washed after running and then incubated overnight in an assay buffer containing 10 mM Hepes, pH 7.6, 20 mM cellobiose, 8 mM MgCl2, 1 mM CaCl2, 1 mM UDP-glucose, 0.1 mM c-3',5'-GMP, and 0.02 % digitonin. Tinopal was used to stain the b-glucan products on the native gels. Gels were photographed under long wave UV-light after washing the stained gels with ethanol solution.
RESULTS
Figure 1. EDTA and EGTA effects on the preparation and solubilization
of the cotton fiber membrane proteins. 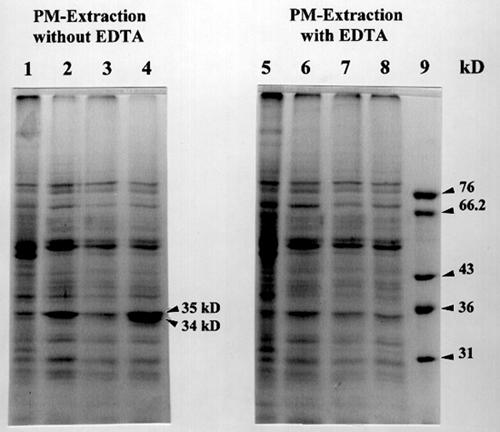
Left panel: Lane 1, the membrane proteins extracted without EDTA.
Lane 2, 10 mM EGTA-wash of membrane fraction (lane 1).
Lane 3, solubilized proteins from 10 mM EGTA-washed membrane.
Lane 4, solubilized proteins from lane 1 fraction with digitonin.
Right panel: Lane 5, the membrane proteins extracted with EDTA.
Lane 6, 10 mM EGTA-wash of membrane fraction (lane 5).
Lane 7, solubilized proteins from 10 mM EGTA-washed membrane.
Lane 8, solubilized proteins from lane 5 fraction with digitonin.
Figure 2. Ca2+ and Mg2+ effects on the product entrapment reaction
analyzed by SDS-PAGE and activity assay.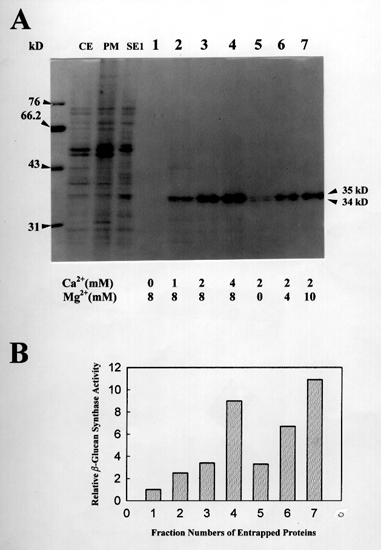
(A) CE, crude extracts of cotton fibers; PM, plasma membrane proteins; SE1, 0.05 % digitonin-solubilized enzymes from PM. Lanes 1 to 7, the entrapped proteins after product entrapment of SE1 fraction; the entrapment reactions were performed under different Ca2+ and Mg2+ concentrations as described below the gel. Note that the entrapment of 34 and 35 kD polypeptides was affected by both Ca2+ and Mg2+ concentrations in the reactions.
(B) Relative b-glucan synthase activities of the entrapped proteins. (The fraction numbers in B match with the lane numbers in A). Note that the relative b-glucan synthase activities seem to reflect the relative quantity of 34 and 35 kD polypeptides in the gel.
Figure 3. Photoaffinity labeling with azido-UDP[P32]-glucose.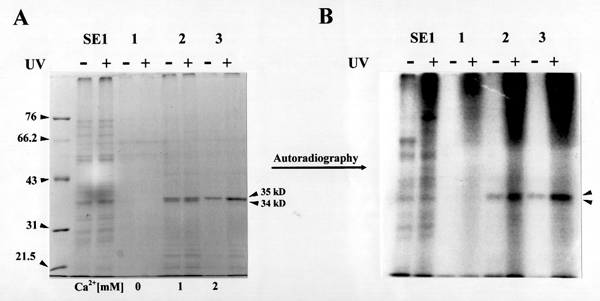
Enzyme fractions were incubated in b-glucan synthesis reaction mixtures with azido-UDP[P32]-glucose in the presence (+) or absence (-) of UV. After reactions, the proteins were collected and separated by SDS-polyacrylamide gel electrophoresis.
(A) SDS-denatured gel stained with Coommassie-blue. SE1 : solubilized enzymes with 0.05 % digitonin. Lane 1 : Product entrapment of SE1 in the absence of Ca2+. Lane 2 : Product entrapment of SE1 in the presence of 1 mM Ca2+. Lane 3 : Product entrapment of SE1 in the presence of 2 mM Ca2+.
(B) Autoradiograph of the same gel. Notice the 34 and 35kD bands which bind UDP-glucose as a function of protein quantity.
Figure 4. Peptide Mapping with V8 Protease 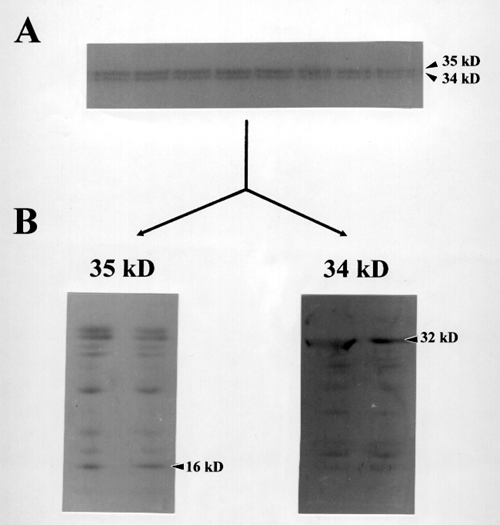
(A) Preparative SDS-polyacrylamide gel electrophoresis. 10 % acrylamide in running gel was used to separate proteins after product entrapment reactions. The 34 and 35 kD bands were cut out after electrophoresis and subjected to proteolytic digestion with V8 protease to get N-terminal amino acid sequences of internal peptides.
(B) Cleveland maps of cotton 34 and35 kD polypeptides
V8 protease-digested proteins were electroblotted to a PVDF membrane and stained with Coomassie brilliant blue. Pannel 34 and 35 kD show Cleveland maps of 34 and 35 kD polypeptides respectively.
Figure 5. Alignment of partial amino acid sequence of cotton
fiber 35 kD polypeptide with other plant and animal derived annexin
sequences.
Figure 6. Preparative native gel electrophoresis of digitonin-solubilized
cotton plasma membrane proteins. 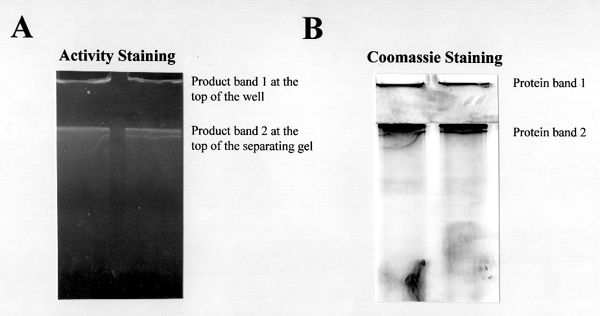
(A) In situ b-glucan synthase activity assay: The gel after electrophoresis was treated with UDP-glucose plus b-glucan synthase effectors for overnight and the b-glucan synthase activities were identified in situ by staining the b-glucan products with calcofluor dye. (The b-glucan products in the gel were subjected to product analysis by transmission electron microscope)
(B) Coomassie-blue-stain of the same gel.
Figure 7. Electron microscopic analysis of product band 1 from
native gel. 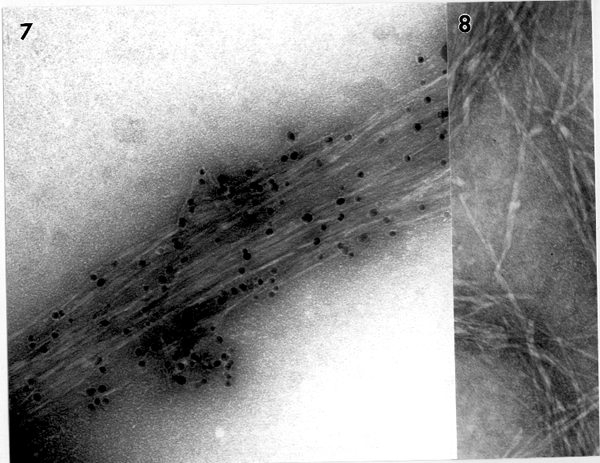
Figure 8. Electron microscopic analysis of product band 2 from native gel. The product band 2 from the native gel of figure 6 was negatively stained and CBH I-gold-labeled. By TEM observation, typical b-1,3-glucan structures were found to be abundant in this band.
DISCUSSION
Based on our data of photoaffinity labeling experiment and product entrapment, we would like to propose new biochemical functions for cotton annexin-like proteins in b-glucan biosynthesis. As recently suggested by Andrawis et al, at a first glance cotton annexin-like proteins appear to negatively affect callose synthase activities in cotton membranes. Membrane-washing with 5 mM EDTA greatly helped to remove the annexin-like proteins, 34 and 35 kD, from cotton membranes (Figure 1), which resulted in total b-glucan synthase activity increase (Table 1). By the way, the relative total b-glucan synthase activities seem to reflect the relative quantity of entrapped annexin-like proteins (Figure 2), indicating a positive regulation of callose synthase activity by annexin-like proteins. So it is still obscure to us how cotton annexin-like proteins are involved in b-glucan biosynthesis. Interestingly, the 34 and 35 kD bind azido-UDP-glucose as a function of protein quantity (Figure 3), maybe suggesting annexin-like proteins as UDP-glucose transporters for b-glucan synthesis. As UDP-glucose transporters, the cotton annexin-like proteins might be able to regulate b-glucan biosynthesis by regulating substrate concentrations in cotton fiber cells. More biochemical experiments are in progress to confirm our idea of cotton annexins as UDP-glucose transporters in b-glucan biosynthesis.
Using TEM analysis, we identified in vitro b-glucan products synthesized in native gels. Preliminary data show that enzyme complexes responsible for some b-glucan synthesis are too big to get into the gel (see product band 1 in Figure 6A), and only celllulose is seen by TEM when analyzing the b-glucan product bands at the wells of gel (Figure 7). More biochemical experiments are under progress to support this new discovery of a cellulose synthesizing complex using native gel electrophoresis.
REFERENCES
1. Geisow, M.J., Walker J.H. (1986) Trends Biochem Sci. 11, 420-423.
2. Burgoyne R.D., Geisow M.J. (1989) Cell Calcium. 10, 1-10
3. Creutz, C.E. (1992) Science. 258, 924-931.
4. Pirck, M., Hirt, H., and Herberle-Bors, E. (1994) Plant Physiol. 104, 1463-1464
5. Boustead, C.M., Smallwood, M., Small, H., Bowles, D.J. and Walker, J.H. (1989) FEBS Lett. 244, 456-460.
6. Blackbourn, H.D., Barker, P.J., Huskisson, N.S. and Battey, N.H. (1992) Plant Physiol. 99, 864-871.
7. Andrawis, A., Solomon, M. and Delmer, D.P. (1993) The Plant Journal. 3(6), 763-772.
8. Kang, M.S., Elango, N., Mattie, E., Au-Young, J., Robbins,P. and Cabib, E. (1984) J. Biol. Chem. 259, 14966-14972.
9. Lin, F.C., Brown, R.M., Jr., Drake, R., Jr. and Haley, B.E. (1990) J. Biol. Chem. 265, 4782-4784.
10. Dhugga, K.S., Ray, P.M. (1994) Eur. J. Biochem. 220, 943-953.
11. Okuda, K., Li, L., Kudlicka, K., Kuga, S. and Brown, R.M.,Jr. (1993) Physiol. 101, 1131-1142.
12. Krystyna, K., Brown, R.M., Jr., Li, L., Lee, J.H., Shin, H. and Kuga, S. (1995) Plant Physiol. 107; 111-123.
13.Cleveland, D.W. (1983) Methods Enzymol. 96, 222-229.
14. Matsudaira, P. (1987) J. Biol. Chem. 262, 10035-10038.